Metric Generation and Analysis Suite (M.G.A.S.)
See the associated GitHub repo for more details, and to submit issues, here
A set of Python scripts I developed to help me analyze large datasets of face-embedding or style distance data, developing a primitive Generalized Accuracy Score (GAS) based on various statistical methods. The intention is to compare two, or more, GenAI models (e.g. Stable Diffusion; trained on the same subject or style) by sheer BRUTE FORCE, comparing ensembles of randomly generated images from each against a fixed set of subject reference images using various statistical metrics/weights.
Such a comparison can be crucial in determining the effect of a single change in training, or image generation, hyperparameters and its overall effect on model output. For example, seeing the effect of changing the Loss Weight function between 'Constant' and 'Min_SNR_Gamma', or seeing the difference between the image samplers 'DPM++2M' and 'DPM++2M-SDE', on what is otherwise the same dataset and hyperparameters. Use it to check that sketchy claim that your friend's favorite custom sampler is the "best" or is "way better" than DPM++2M-Karras.
Ultimately, the success of a realistic or style model is determined by its ability to reliably reproduce the likeness of the trained subject material, and this script is my first serious attempt to quantitatively address issues related to that. With the addition of the GUI and CREATE scripts, this suite endeavors to give the community an easy, standardized way to approach model training and testing.
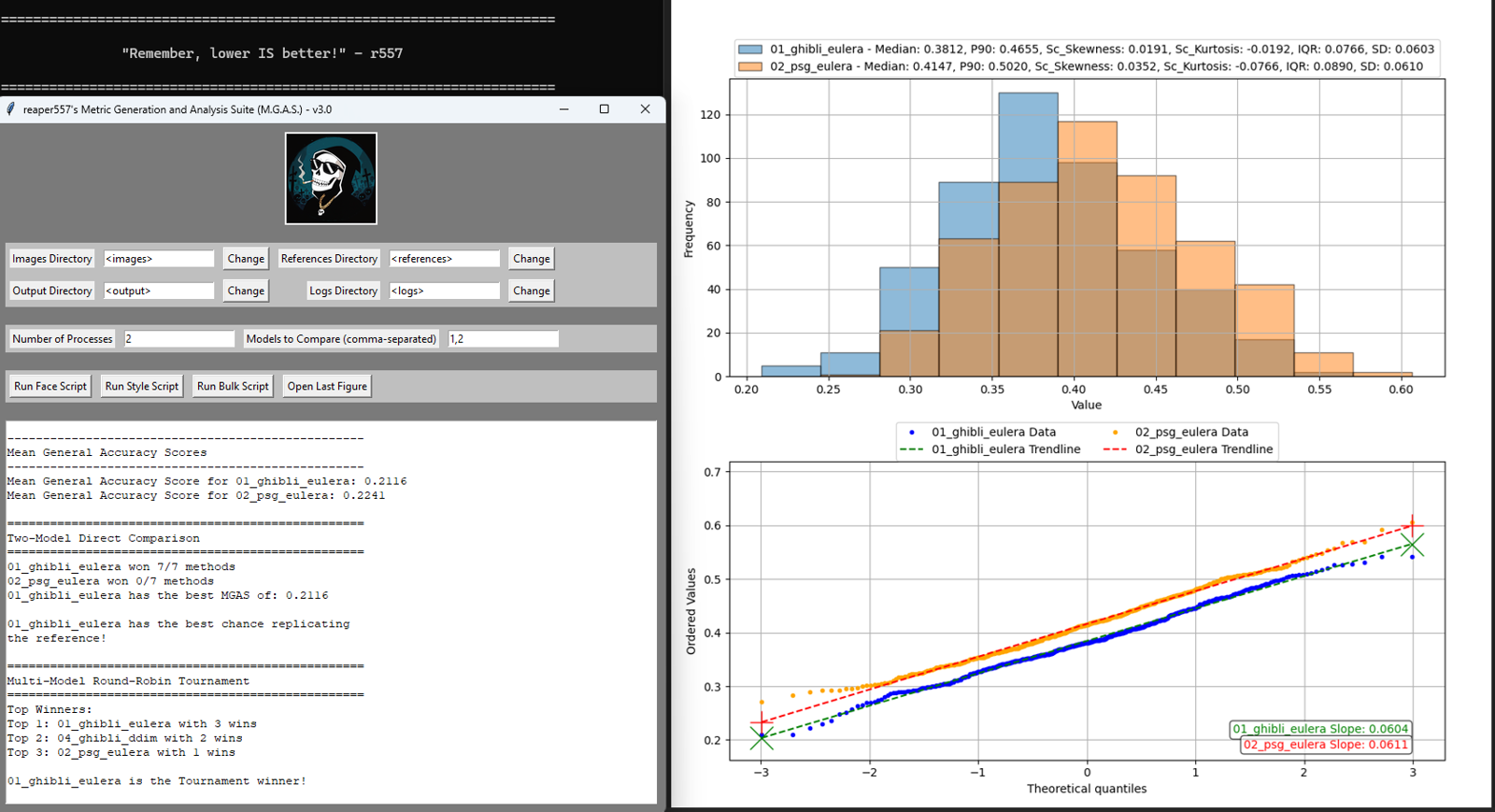
Ever thought one model was more often accurate, but this other one is a bit more flexible? You can see this immediately in the figure's histogram. Wondered if your regularization (REG) set was large enough to properly balance your image set? Compare it to your image data set and see any significant correlation through the Q-Q plot; good sets have nearly straight lines while imperfect sets have curled ends.
This GUI app now has the ability to create the distance data in the required list format on its own through the CREATE script; no external nodes/extensions/apps needed to use my scripts!
Built with the help of GPT-4o, and that's The Toasted Toad's Truth, thanks Zedd! (>'.')>[<3]
Feel free to use, share, and modify these scripts to suit your needs. (MIT - license)
Made "for fun" and shared completely for free to those who love GenAI.
<(˶ᵔᵕᵔ˶)>
Changelog
[3.0] - 2025/04/09
Major update that adds the ability to test style models for analysis with the bulk script.
Added
Added
create_styledata.py– evaluates style similarity between reference and generated images using a CLIP ONNX model.Added
onnxruntime==1.17.0to support ONNX inference
Changed
The
install_MGAS.batnow checks for the new model,clip-ViT-B-32-vision.onnxas well, which is hosted by Qdrant on HuggingFace.Renamed “Run Create Script” button to “Run Face Script” for clarity alongside the new “Run Style Script”.
The venv setup now expects Python 3.10 to be installed beforehand with a preference for
3.10.11.
Fixed
Fixed the face and bulk scripts not properly using gui path enviroment variables (Oops!).
[2.6] - 2024/12/22
Similar to the previous update, this one extends the facial recognition workflow to the generated image folders (Oops!), further optimizes the multithreading method with batches, and improves logging and error handling.
Added:
Added functionality to validate reference embeddings during runtime.
Added functionality to save reference embeddings in
CACHEfolder to save time with repeated use.Enhanced logging levels (DEBUG, INFO, WARNING, ERROR, CRITICAL) to allow more detailed debugging and runtime insights. Advanced users can set the
LOG_LEVEL(e.g. "DEBUG") at the top of the scripts for more or less information in the logs.Implemented a robust file integrity check for output data, ensuring that files contain valid, parsable data before proceeding with analysis.
Added functionality to detect and handle corrupted files gracefully by skipping them and logging detailed error messages in a new file,
failed_files.txt.
Changed:
Updated multiprocessing to utilize Pool.imap_unordered, enabling more efficient distribution of tasks across available processes.
Updated tasks to be set up using dynamic batch sizes tied to the user defined
num_processes.Restructured the image embedding computation process to reduce redundancy and improve readability.
Improved histogram generation with consistent bin sizes for better visual comparison between datasets.
Fixed:
Fixed the alignment and preprocessing steps not being applied to image folders (Oops!).
Fixed
install_MGAS.batsuch that it creates all needed directories for the user (no longer need to DELETE TXTs).Fixed a bug in handling output files with invalid or missing data.
Improved error handling for missing or incompatible DLib models.
[2.5] - 2024/12/19
This update focuses on properly implementing the facial recognition workflow from the methodology section and optimizing the parallel processing to increase speed.
Added:
Implemented the Alignment and Normalization steps through the
align_facefunction along with DLib'sget_face_chip. Faces are now properly preprocessed before the embeddings are calculated.Included the 68-point landmark model for use in the Alignment step and increased accuracy. This now replaces the previously used 5-point model in all use cases.
Fixed:
Fixed the absolute mess that the parallel processing functionalities in the
process_imagesfunction loop were. This has significantly increased processing speed and improved CPU usage.Fixed the BULK script to use alphanumerical sorting rather than lexicographical.
Improved logging and error reporting relevant to the
process_log.txtfile.
[2.4] - 2024/12/09
After taking a long break, I found some gnarly issues while testing a new model under relatively fresh installs of Python/SwarmUI/etc.
Added:
Included the
HF_model_download.pyto handle downloading the two face-recognition DAT files from HuggingFace. This now also replaces the janky curl method in theinstall_MGAS.batAdded huggingface_hub to the
requirements.txtAdded a Methodology section to the GitHub repo for clarity.
Fixed:
Fixed how the scripts were calling the local venv. Although the venv is activated when the GUI starts, the
gui.pyscript itself was not enforcing this when calling the other scripts. Thegui.pyscript now properly ensures that the venv python is used in therun_scriptmethod.Updated the Installation and Usage section for clarity.
[2.3] - 2024/07/31
Changed:
Changed how model names were assigned. Now, model names follow their folder names instead of the generic 'model_x' format, and this change should be reflected in the corresponding LOGS.
Changed how model names were sorted regarding the 'models to compare directly' variable. Now, model order is prepended onto folder names such that which two models are chosen is no longer ambiguous, e.g. 'model_name' --> '1_model_name'. Python was sorting by name in a slightly different way from Windows, and this was resulting in the incorrect folders being assigned to the 'models to compare directly' variable; i.e. script order was not matching what the user could see in the 'output' folder itself.
[2.2] - 2024/07/30
Changed:
Changed how the results of the Round-Robin were determined and reported. The convoluted MGAS system has been replaced with a simple, but effective, tally system.
Results of the Round-Robin now "make sense" and should now accurately reflect the results from Two-Model Direct Comparisons.
Changed Round-Robin results to only show results of top three winners.
Changed the GAS back to its intended definition, that of a score determined by an individual weight method.
Changed the MGAS back to its intended definition, that of a simple Mean-GAS for each model based on all weight methods.
[2.1] - 2024/07/29
Added:
Included the
optimize_facedata_weights.pyused to generated associated weights in_EXTRASfor user referenceIncluded additional experimental data in
Example_Data_Directoriesover extracted LoRA Rank(Dim) and Alpha combinations, as well as updated example figures and logsAdded
Weighted Rank Sum-Based Weights to list of used methodsAdded my personal
Meat-N-Potatoes-Based Weights as a "sanity check"
Changed:
Changed the previous 'Subjective' Weights to the
OptimizedWeights based on included optimization scheme and new order of import of the metricsChanged the PCA method to the
Robust Principal Component Analysis-Based Weights to better handle noisy dataChanged the Kurtosis and Skewness to,
Sc_Kurtosis&Sc_Skewness, which are scaled versions that have the correct "units" as the data
Fixed:
¡HUGE! Metric redundancies were curtailed through correlation analysis, and a minimum set of metrics was chosen through optimization
¡HUGE! All metrics have been scaled and standardized to ensure that they have the same "units" as each other and the data
¡HUGE! GAS/MGAS calculations fixed to better capture the notion/maxim that, 'more overall low-valued data, the better the distribution'
Fixed some GUI options not giving the appropriate warning when the user puts in inappropriate values; 'Number of Processes' & 'Models to Compare'
Fixed hyphen in GUI start message that was "too dang close"
[2.0] - 2024/07/26
Added:
New
create_facedistance_data.pyscript to carry out data generation without the need for an external app.New
gui.pyscript for GUI interface; userun_GUI.batto begin app and use both main scripts.Added
Multi-Model Round-Robintournament style comparison capability which allows for more than two models to be compared at once (old way is now called Two-Model Direct Comparison).Added the
Kolmogorov-Smirnovnormality test; code will switch from theShapiro-Wilktest when data sets get sufficiently large (~5000; see logs for more info).Added
install_MGAS.batto create venv with required Python dependencies and to automatically download DLib models (from HuggingFace, thanks cubiq!) needed for the CREATE script intoDLIBfolder.Added additional information via two
PLEASE_READ_ME.txtfiles, one in main and other in example directories.
Changed:
Reordered the
order_of_import_of_metricsvariable in the BULK script.Changed what information is made immediately available, via terminal/GUI; check
LOGSdirectory for detailed logs.Directory path assignment, and important user defined variables, now accessed through GUI; no longer need to edit any of the scripts!
[1.0] - 2024/07/23
First release.
Installation
1. Please have Python 3.10 installed (use 3.10.11 for best results). You can download it from Download Python in the "Looking for a specific release?" section by scrolling down (3.10.11 at release date, April 5, 2023).
2. Clone the repository and go into the created folder:
git clone https://github.com/klromans557/Metric-Generation-and-Analysis-Suite
cd Metric-Generation-and-Analysis-SuiteOR
Download the attached zipped repository files
3. Install the required dependencies (txt file in zip):
Use the supplied
install_MGAS.batfile.Installs the empty directories and model files (~ 400MB) too!
Usage
To use the script and analyze the face-distance data, follow these steps (also see PLEASE_READ_ME.txt ([WIP] outdated) file in the main repo directory for more details):
Use the provided
run_GUI.batfile to open the GUI and run the scripts.Make sure your image folders and fixed set of reference images are in the correct directories within
DIRbefore clicking theRun Face ScriptorRun Style Scriptbutton. These scripts will generate the face or style similarity metric data in theoutputfolder.Within
imagesfolder: place folders of images that will represent "models" to be tested. For example, a folder called "dpmpp" and another called "euler", each filled with 10 images generated using those respective samplers (all other parameters fixed).Within
referencesfolder: place a fixed set of reference images to compare the "models" to. Data for each reference is collected together for the corresponding tested model. For example, use 5 images of your main subject here. Each model will then get 10x5=50 data points for the BULK statistics. Results improve with more images/refs!
Data created, or placed, into the
outputfolder is accessible to the BULK script. UseRun Bulk Scriptbutton to perform statistics on data.Data on the metrics/weights, results of the comparisons, and the tournament winner will then be displayed in the GUI text box.
All created data, including the normality tests not usually shown, are stored in the
LOGSdirectory.
Press the
Open Last Saved Figurebutton to open the last figure made by the BULK script. Note that any changes will require the BULK script to be re-run to update the figure.Use the
Models to CompareGUI variable to change which two models are plotted in the figure and used in the Two-Model Direct Comparison; models are ordered by name, ascending, i.e. the default '1,2' compares the first and second model folders.Use the
Number of ProcessesGUI variable to change how many CPU cores/threads are used for processing images with the CREATE script (default '2' works well; be careful not to set this too high, e.g. ~10 for an i5-14600K).
[EXAMPLES] See the second
PLEASE_READ_ME.txt([WIP] outdated) in the example data directory, and associated folders, to get an idea of the potential uses for the script. Included are some details on four experiments I ran using M.G.A.S.
Methodology
Facial Recognition
The heart of the method relies on the similarity of facial embedding distance data created by the face recognition models. Roughly speaking, the model finds 68 landmarks on the reference's face, i.e. eyes, nose tip, mouth corners, etc., and assigns a distance value (e.g. 0.3). The process unfolds as:
1. Face Detection: The model first identifies the general region of the face, just like a bounding box.
2. Landmark Detection: The landmarks are identified within the face region.
3. Alignment: The face is then "aligned" using the landmarks. This step accounts for various poses by adjusting faces to a standard forward-facing form. In other words, a tilted face is adjusted such that the eyes are horizontal and the face upright.
4. Normalization: The aligned face is cropped & resized to fit a model-defined standard size (e.g. 150x150 pixels). This ensures uniform processing.
5. Feature Extraction: The normalized face is then analyzed for unique features. These features then form a kind of "fingerprint" of the face which can be used for recognition.
6. Recognition: Finally, the extracted features are compared to a known database of faces. In the case of MGAS, this database comprises the reference images placed by the user. The system analyzes how similar the faces are and generates a distance value based on this association (e.g. 0.3). The lower the value the closer the tested image matches a given reference. This is the origin of the maxim, "Lower is better!"
From a given set of images to test and another set of reference images to form the database, a large set of statistical data can be generated by these similarity measurements.
Style
[WIP]
Metrics
Once the base statistical data is generated it can be analyzed with various metrics (see the main repo image for an example of a histogram created from such data). I chose a set of six metrics, i. Median, ii. 90th-Percentile (P90), iii. Scaled Skewness, iv. Scaled Kurtosis, v. Interquartile range (IQR), and the Standard Deviation (SD), with the order set by importance. This set was chosen to best minimize redundant correlations between the metrics and to maximize accounting for distribution shape and position. A "good" distribution is not only centered on a low value (i.e. low median), but does not tend to produce high-valued outliers (i.e. skewness < 0):
1. Median: This is the middle value of the dataset when the set is ordered. This behaves effectively the same as the Mean (or Average) for the kind of data we will analyze with MGAS, i.e. represents the center of the histogram, but it is less susceptible to extreme values and outliers. For example, when viewing the main repo histograms, the median is around 0.33 which corresponds to the point on the x-axis under the highest part of the distribution.
2. 90th-Percentile (P90): Is the value below which 90% of the data points fall, or in other words, the highest 10% of the data is beyond this point. This value helps describe the tail of the distribution and gives an idea of the magnitude of the higher-end values.
3. Scaled Skewness: Skewness measures the symmetry of the distribution. For a perfectly symmetric distribution, like the Gaussian, this metric is 0. Positive Skewness values correspond to a longer right-side tail, while negative values are for a longer left-side tail. I have scaled these values to ensure they have the same "units" as the other metrics to keep comparisons meaningful.
4. Scaled Kurtosis: Regular Kurtosis (or sometimes Excess Kurtosis) measures the "tailedness" of a distribution or how extreme the outliers are. A Gaussian distribution has a Kurtosis of 3 (Excess Kurtosis of 0), meaning it has a moderate tail length. Positive Kurtosis has "fat"-tails, a sharp peak, and a higher presence of extreme values, while negative values correspond to the opposite. I have scaled these values to ensure they have the same "units" as the other metrics to keep comparisons meaningful.
5. Interquartile Range (IQR): This is the range of values between the 25th and 75th percentiles. It focuses on the spread of the middle 50% of the data, ignoring the tails, and gives a sense of distribution variability and data compactness.
6. Standard Deviation (SD): This is the measure of the average spread of data points around the mean. It also gives a sense of distribution variability taking into account the tails.
Weights
With the metrics at hand, we can now use them in various statistical weighting schemes, i.e. different types of averages. Using more than one method leverages the advantages/disadvantages of different distributions without shoe-horning a "one-size-fits-all" solution. Each method will generate an individual GAS score:
1. Uniform Weights: Each metric is treated equally with the same weight value (1/6 ~ 0.167) assigned to all. This is the simplest approach as it is just a basic average.
2. Optimized Weights: These weights were calculated based on the order of import of the metrics and to reduce redundancy from metrics that contain similar information. The script used to optimize is located in the _EXTRAS directory.
3. Weighted Rank Sum Weights: Metrics are ranked based on their values for each dataset, and then the ranks are weighted. This scheme emphasizes the relative position of metrics rather than raw values, making it less sensitive to extreme values and outliers.
4. Inverse Variance Sum: Metrics with lower variability (variance) are given higher weights since they are considered more reliable. This gives stable metrics a larger impact on the analysis.
5. Analytic Hierarchy Process weights: A structured method where metrics are compared pairwise to determine their relative importance.
6. Robust Principal Component Weights: This statistical technique focuses on combinations of metrics that cover the most variation in the data. The weights are derived from these combinations, focusing on capturing the key features of the dataset while reducing noise.
7. "Meat-N-Potatoes" Weights: My sanity check weights which only use the Median and P90 weighted equally (1/2 = 0.5).
After all the weights are calculated, the final MGAS score is found by a simple average, or mean, of the seven weight scheme values.
Direct Comparisons and Round-Robin Tournament
1. Two-Model Direct Comparison: The two models from the Models to Compare GUI variable will be compared, and their respective GAS and MGAS scores calculated. The model which won the most weight scheme comparisons and has the lowest MGAS value is declared the winner.
2. Multi-Model Round-Robin Tournament: All models used by MGAS will be compared here by pairwise direct comparison competitions. The model that wins the most comparisons is declared the winner, with the top three listed in the GUI as well.
Acknowledgments
Matteo Spinelli [a.k.a. cubiq] for creating the ComfyUI custom node that I used as inspiration for the CREATE script and for hosting the DLib model files on HuggingFace.
Qdrant for converting and hosting the CLIP ONNX model on HuggingFace.
OpenAI & Alibaba Cloud for providing guidance and assistance in developing this project.
GitHub for hosting the repository.
Dr. Furkan Gözükara for sharing his scripts through the SECourses Civitai, Patreon, and the associated Discord server and YouTube channel. These resources were invaluable to me during the development of this project and served as guides/templates for creating such scripts.